
Nucleic acids such as DNA and RNA exhibit a wide range of biochemical functions including transfer of genetic information, regulation of gene expression, molecular recognition, and chemical catalysis. The objective of Nucleic Acid Chemistry and Engineering Unit is to harness the intrinsic functional versatility of natural and artificial nucleic acids to engineer nucleic acids with sophisticated functions.
Actual projects that postdocs and students work on largely depend on individual background and interests within the context of nucleic acid chemistry and engineering. For example, we currently have a variety of projects such as:
- Applications of high-throughput sequencing to analyze and engineer nucleic acid enzymes.
- Development and applications of riboswitches in mammalian cells, viruses, bacteria, and artificial cells.
- DNA-programmed self-assembly.
Our postdocs and students come from a variety of backgrounds in chemistry and life sciences.
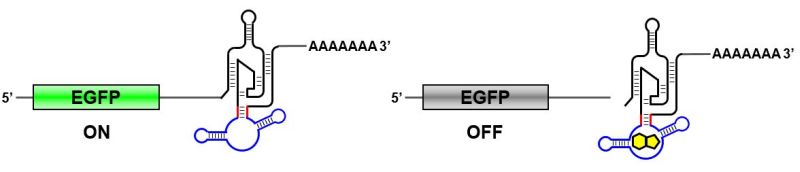
Engineered nucleic acids, RNA in particular, can potentially be used within living cells to interact with the intracellular molecules and control how cells behave. Over the years, we have designed various RNA-based gene switches (riboswitches) that can regulate gene expression in bacteria and in mammalian cells in response to chemical signals. We are also interested in exploiting these synthetic RNA devices for other applications such as metabolic engineering, stem cell engineering, and gene therapy. Several collaboration projects with industry and academic partners are ongoing. For these types of projects, we look for researchers with diverse life sciences background who can contribute ideas and skills for new applications.
We are interested in designing simple and complex chemical systems composed of multiple nucleic acid components and modules. We recently designed an RNA biosensor circuit that amplifies a chemical signal and produces an optical signal (fluorescence). These synthetic chemical systems composed of nucleic acids not only challenge the limits of nucleic acid chemistry, but also serve as models for future biological applications.
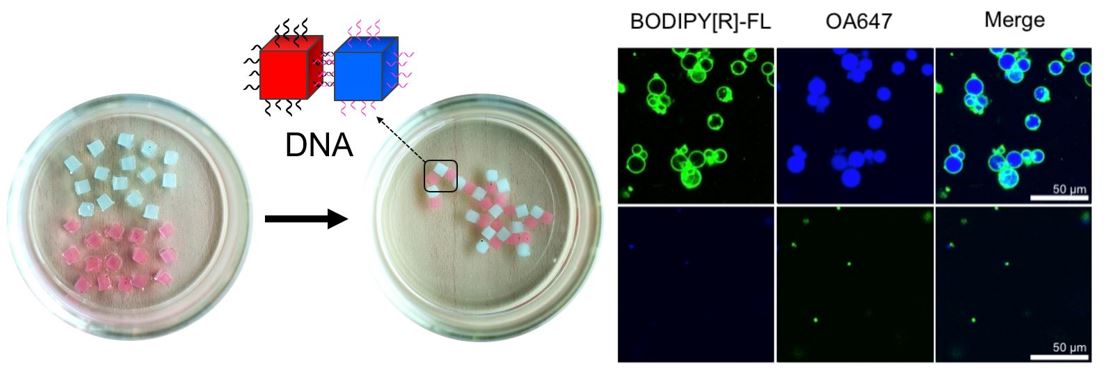
We are also interested in using unnatural nucleic acids to expand the scope of nucleic acid-based chemical systems, macroscopic self-assembly, developing novel aptamers for biological applications, developing novel small molecules to control nucleic acids, constructing artificial cells that encapsulate nucleic acid devices, etc. For these efforts, we welcome nucleic acid chemists with organic synthesis skills.
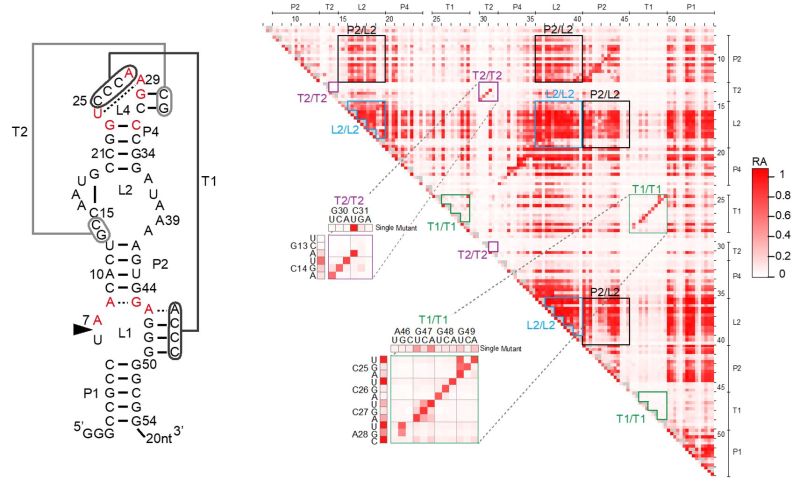
Development of new or improved methodologies and techniques is often necessary to design nucleic acids with sophisticated functions. We have developed and will continue to develop new methods that significantly accelerate our ability to design and analyze functional nucleic acids. These methods are used, along with other available methods, for various applications.
For example, we are developing new methods to use high-throughput sequencing to quantitatively analyze tens of thousands of ribozyme and deoxyribozyme mutants. We use this method to engineer better riboswitches that control gene expression in living cells as well as to better understand the sequence-function relationships of nucleic acid enzymes. We are also adapting microfluidic technology to rapidly engineer other types of nucleic acid devices. We recently developed a new selection method to engineer specific RNA-protein binding pairs.
